Fig. 3
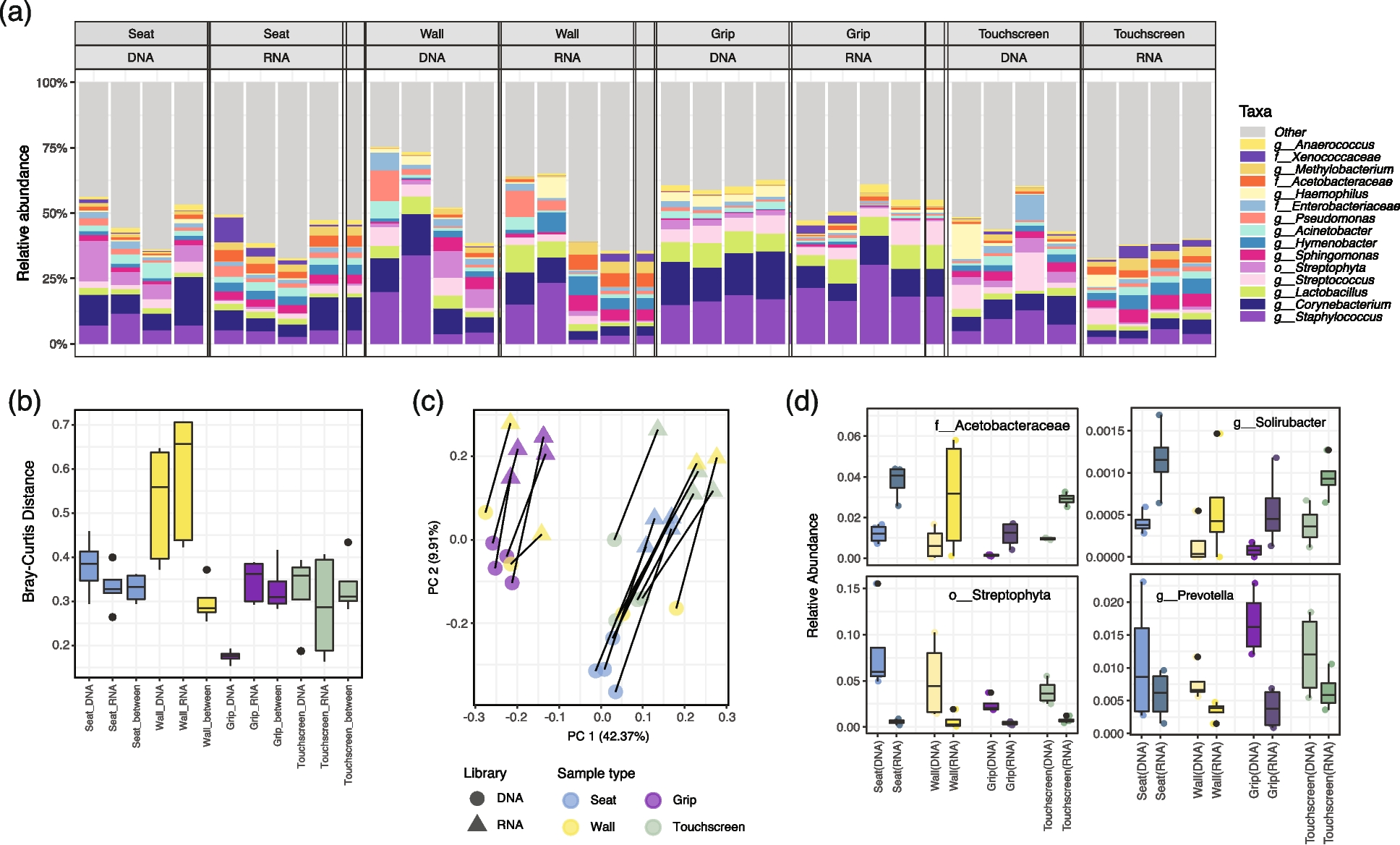
16S-RNA-seq indicated subtle differentiation between DNA and RNA libraries in samples from the Boston (MBTA) subway system. a Relative abundances of the 15 taxa with the highest means across four sample types in DNA and RNA libraries indicated overall similar taxonomic compositions. Each column represents a biological replicate. b Bray–Curtis distance distributions between MBTA samples within/between DNA and RNA libraries. c Principal coordinate analysis (PCoA) of MBTA samples using Bray–Curtis distances. Sample type and library type are both drivers of overall community composition. d Four differentially abundant taxa that consistently enriched or depleted in RNA libraries across sample types. The Acetobacteraceae family and Solirubacter genus were consistently enriched in RNA libraries, while the Streptophyta order and the human commensal Prevotella genus were under quantified in RNA libraries (mixed-effects linear models, q-value = 0.002, 0.003, 0.002, and 0.012, respectively)